This webpage was produced as an assignment for Genetics 677, an undergraduate course at the University of Wisconsin - Madison.
DNA Motifs
A motif is a string of nucleotides in a DNA sequence which is repeated , or conserved in all sequences in a multiple alignment at a particular position . Motifs may correspond to structural or functional elements within the sequence they characterize.
MOTIF
No motifs were found using MOTIF (1), because the nucleotide sequence is so large. Therefore, I used the intron-free CFTR mRNA sequence. Using mRNA, many motifs were found, depending upon the cut-off value used.
Cut-off value: 85
Number of found motifs: 161
Cut-off value: 95
Number of found motifs: 28
Cut-off value: 97
Number of found motifs: 15
Cut-off value: 100
Number of found motifs: 11
Cut-off value: 85
Number of found motifs: 161
Cut-off value: 95
Number of found motifs: 28
Cut-off value: 97
Number of found motifs: 15
Cut-off value: 100
Number of found motifs: 11
MOTIF returned 11 motifs, at a cut-off value of 100, of the mRNA of CFTR:
Motif Name Sequence Number Function
1. HSF AGAAN 2 Heat Shock Factor
2. ADR1 NGGRGK 1 Alcohol Dehydrogenase Gene Regulator 1
3. CdxA MTTTATR 2 May be part of network regulating gastrulation
4. NIT2 TATCTM 1 Activation of nitrogen-regulated genes
5. SRY AAACWAM 1 Sex-determining region Y gene product
6. STRE TMAGGGGN 1 Stress response element
7. AML-1a TGTGGT 1 Runt-factor AML-1
8.GAmyb YAACSGMC 1 GA-regulated myb gene from barley
9. GATA-1 NCWGATACA 1 GATA-binding factor 1
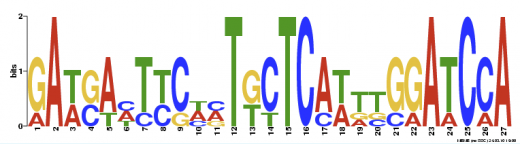
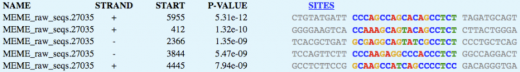
MEME
MEME (2) only allows nucleotide sequences of 60,000 base pairs or less. The CFTR sequence was too large for the search engine and the mRNA sequence was used instead. Three motifs were found in the sequence.
Motif 1
Motif 2
Motif 3
Analysis
Using MOTIF, only 11 motifs were identified at the 100 cut-off value, indicating that these motifs are truly found within the CFTR mRNA. MOTIF clearly states the names and sequences of motifs, while MEME gives a more aesthetic view of each motif sequence found. However, MEME did not give the name of the sequence or its function.
_______________
References
1. MOTIF. March 8, 2010. http://motif.genome.jp/
2. MEME. March 11, 2010. http://meme.sdsc.edu/meme4/intro.html
3. NCBI: Entrez Nucleotide. "CFTR mRNA Sequence". March, 11, 2010. http://www.ncbi.nlm.nih.gov/nuccore/NM_000492?report=fasta&log$=seqview
References
1. MOTIF. March 8, 2010. http://motif.genome.jp/
2. MEME. March 11, 2010. http://meme.sdsc.edu/meme4/intro.html
3. NCBI: Entrez Nucleotide. "CFTR mRNA Sequence". March, 11, 2010. http://www.ncbi.nlm.nih.gov/nuccore/NM_000492?report=fasta&log$=seqview
Alexandra Reynolds
[email protected]